About
| ||
Napolitano, F., Carrella, D., Mandriani, B., Pisonero-Vaquero, S., Sirci, F., Medina, D.L., Brunetti-Pierri, N., and di Bernardo, D. Gene2drug: a computational tool for pathway-based rational drug repositioning. Bioinformatics (2017). Abstract | ||
| ||
|
Drug repositioning has been proposed as a potential effective shortcut to drug discovery. Among the available approaches, rational methods based on a specific molecular target can take advantage of the current availability of genome-wide transcriptional data. Unfortunately, the effects of direct drug targets are usually not detectable at the mRNA level and a more systematic approach is required to link transcriptional data to therapeutic effects. Gene2drug tackles this problem by searching for sets of pathways that are dysregulated by a drug as opposed to single genes. One way to define a set of pathways is to start from a target gene and collect the pathways that it is annotated to, thus assessing the effects of perturbing the target gene indirectly through cellular mechanisms that are expected to be involved. | ||
| ||
|
Gene2drug uses a method we called “Pathway-set Enrichment Analysis” (PSEA), analogous to the Gene-set Enrichment Analysis (GSEA). Gene expression profiles from the Connectivity Map are converted to “pathway expression profiles” and ranked according to the p-value of their Kolmogorov-Smirnov (KS) statistic. Given a set of pathways, the KS statistic is applied again to search for drugs that consistently up-regulate or dysregulate most pathways in the set. | ||
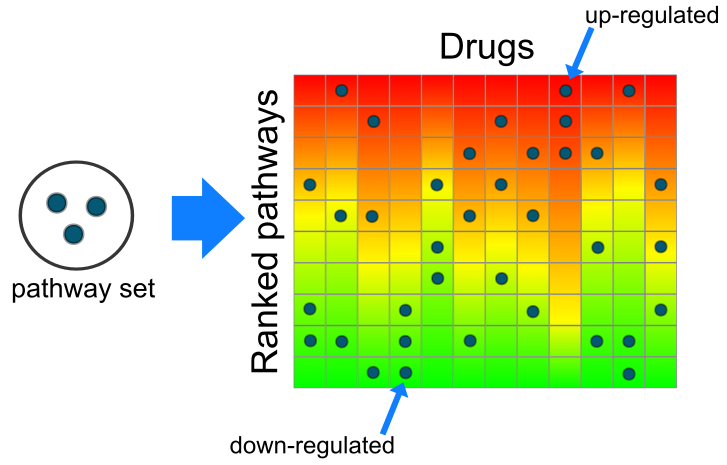
| ||
| ||
| ||
| ||
|
The results page shows a list of (top 10%) drugs as prioritized by the PSEA analysis. Top drugs are those most dysregulating the pathways in the input set. Enrichment score signs report whether the regulation is “up” or “down”. The p-value reports how it is unlikely for a random set of ranks to be as extreme (top or bottom) as the ranks of the chosen pathways for each drug. The “Export Data” button becomes active as soon as a full report of the analysis is ready. The report includes details of the pathway ranks for each drug. Small ranks for a pathway mean that it is upregulated (top of the profile), large ranks mean that it is downregulated (bottom of the profile). The ranks range from 1 to N, where N is the number of database for each profiles (see table above). | ||
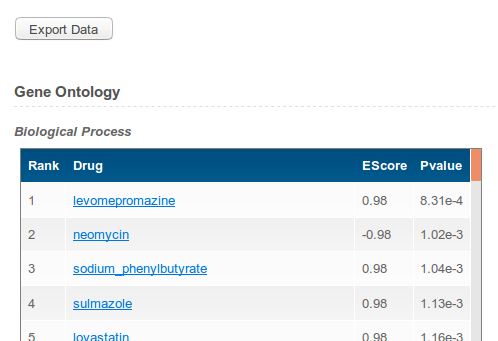
| ||
|
| ||
|
Use of Gene2drug tool is free to academic, government and non-profit users for non-commercial use. Use of Gene2drug application for commercial use by non-profit and for-profit entities requires a license agreement. Parties interested in commercial use may first obtain a demo version of a stand-alone version of Gene2drug application, subject to this License, by contacting Diego di Bernardo (dibernardo_at_tigem.it). Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
|

